Network Hierarchies and the Michelangelo Principle
The Michelangelo Principle — almost certainly apocryphal — states that:
If you want to transform a marble block into David, just chip away the stone that doesn’t look like David.
This seems to be a great idea to solve literally every problem in the world: if you want to fix evil, just remove from the world everything that is evil. But I don’t want to solve every problem in the world. I want to publish papers on network analysis. And so I apply it to network analysis. In this case, I use it to answer the question whether a directed network has a hierarchical structure or not. The result has been published in the paper “Using arborescences to estimate hierarchicalness in directed complex networks“, which appeared last month in PLoS One.

To determine whether a network is hierarchical is useful in a number of applications. Maybe you want to know how resilient your organization is to the removal of key elements in the team. Or you want to prove that a system you mapped does indeed have a head, instead of being a messy blob. Or you want to know whether it is wise to attempt a takedown on the structure. In all these scenarios, you really desire a number between 0 and 1 that tells you how hierarchical the network is. This is the aim of the paper.
Following the Michelangelo Principle, I decide to take the directed network and chip away from it everything that does not look like a perfect hierarchy. Whatever I’m left with is, by definition, a perfect hierarchy. If I’m left with a significant portion of the original network, then it was a pretty darn good hierarchy to begin with. If I’m left with nothing, well, then it wasn’t a hierarchy at all. Easy. Let’s translate this into an algorithm. To do so, we need to answer a couple of questions:
- What’s a “perfect hierarchy”?
- How do we do the chipping?
- How do we quantify the amount we chipped?
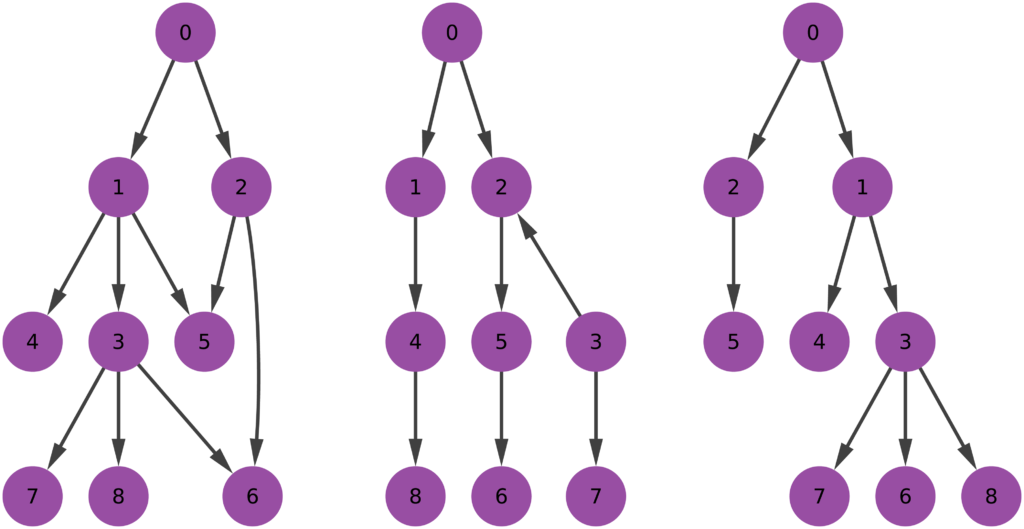
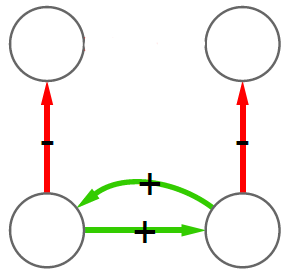
The first question is the one where we have most wiggle room. People might agree or disagree with the definition of perfect hierarchy that I came up with. Which is: a perfect hierarchy is a system where everybody answers to a single boss, except the boss of them all, who answers to no one. I like this definition because it solves a few problems.
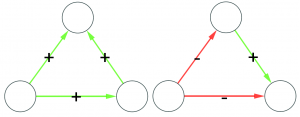
Consider the picture above. In the leftmost example, if we assume nodes 1 and 2 give contradictory orders, node 5 doesn’t really know what to do, and the idea of a hierarchy breaks down. In the example in the middle, we don’t even know who’s the boss of them all: is it node 0 or node 3? The rightmost example leaves us no doubt about who’s boss, and there’s no tension. For those curious, network scientists call that particular topology an “arborescence“, and that’s the reason this exotic word is in the paper title. Since this is a well defined concept, we know exactly what to remove from an arbitrary directed network to make it into an arborescence.
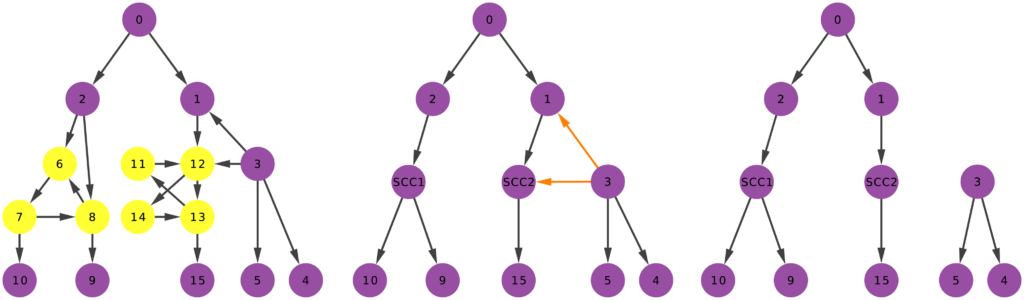
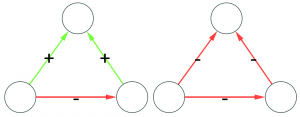
Time to chip! Arbitrary directed networks contain strongly connected components: they have paths that can lead you back to your origin if you follow the edge directions. An arborescence is a directed acyclic graph, meaning that it cannot have such components. So our first step is to collapse them (highlighted in yellow above) into a single node. Think of strongly connected components as headless teams where all the collaborators are at the same level. They are a node in the hierarchy. We don’t care how a node organizes itself internally. As long as it answers to a boss and gives direction to its underlings, it can do it in whichever way it wants.
Second chipping step: in an arborescence, all nodes have at most one incoming connection, and only one node has zero. So we need to remove all offending remaining edges (highlighted in orange above). Once we complete both steps, we end up with an arborescence, and we’re done. (There are edge cases in which you’ll end up with multiple weakly connected components. That’s fine. If you follow the procedure, each of these components is an arborescence. Technically speaking, this is an “arborescence forest”, and it is an acceptable output)
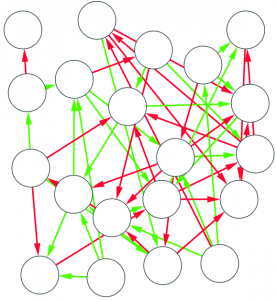
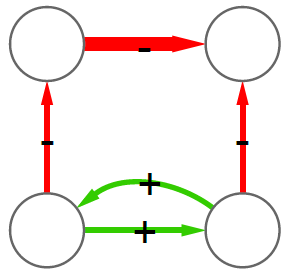
We can now answer the final question: quantifying how much we chipped. I decide to focus on the number of edges removed. Above, you see that the original graph (left) had twenty edges, and that (right) nine edges survived. So the “hierarchicalness” of the original graph is 9 / 20 = .45.
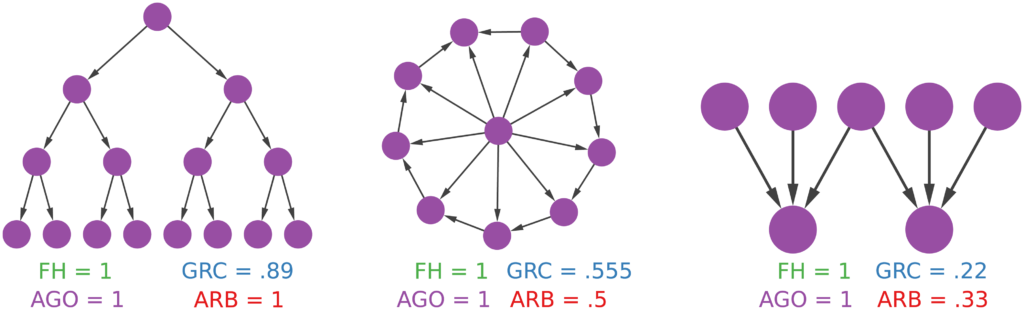
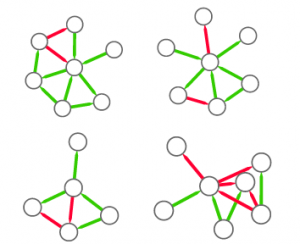
Now the question is: why would someone use this method to estimate a network’s degree of hierarchicalness and not one of the many already proposed in the literature? The other methods all have small downsides. I build some toy examples where I can show that arborescence is the way to go. For instance, you can’t find a more perfect hierarchy than a balanced tree (leftmost example above). However, Global Reach Centrality would fail to give it a perfect score — since it thinks only a star graph is a perfect hierarchy. Agony and Flow Hierarchy aren’t fooled in this case, but give perfect scores in many other scenarios: a wheel graph with a single flipped edge (example in the middle), or a case where there are more bosses than underlings (rightmost example). Those who have been in a team with more bosses than workers know that the arrangement could be described in many ways, but “perfect” ain’t one of them.
Arborescence is also able to better distinguish between purely random graphs and graphs with a hierarchy — such as a preferential attachment with edges going from the older to the newer nodes (details in the paper). Before you start despairing, it’s not that I’m saying we’ve been doing hierarchy detection wrong for all these years. In most real world scenarios, these measures agree. But, when they disagree, arborescence is the one that most often sides with the domain experts, who have well informed opinions whether the system represented by the network should be a hierarchy or not.
To conclude, this method has several advantages over the competitors. It’s intuitive: it doesn’t give perfect ratings to imperfect hierarchies and vice versa. It’s graphic: you can easily picture what’s going on in it, as I did in this blog post. It’s conservative: it doesn’t make the outlandish claim that “everyone before me was a fool”. It’s rich: it gives you not only a score, but also a picture of the hierarchy itself. So… Give it a try! The code is freely available, and it plays nicely with networkx.